In this regard, the efficiency of AI is a tens of thousands of biologists
Author:Institute of Physics of the Ch Time:2022.08.07
In front
At the end of 2021, the "Science" magazine nominated ten annual scientific breakthroughs, and many of them were closely related to life science, and fully demonstrated the strong energy of the cross -collision of life science and other disciplines in the current scientific breakthrough.
Today we will talk about the most attention here, and it is also one for two consecutive years -AI predicts protein structure.

In the past few decades, top structure biologists around the world have completed an analysis of about 180,000 protein structures; but in the past two years, Alphafold has completed the structural prediction of almost all protein in the human body. Why is it so amazing? What are the value?
Let's not talk about the protein structure first. When it comes to artificial intelligence, what do you think of?
I guess most people would think of AlphaGo who defeated humans in Go a few years ago. That game showed the extraordinary ability of artificial intelligence in computing -how to use algorithms to achieve the deduction of Go and better than humans.

I believe many people are still vivid
Then look back at the breakthrough selected by this science, you must have a lot of question marks on your head: what is protein? Is its structure complicated? Why use artificial intelligence to predict the protein structure?
Speaking of protein, I believe that many people are not unfamiliar, and even understand that protein is a "component" that exercise various functions in cells. Not only that, protein is also one of the basic substances for our body. For example, fitness can exercise muscles, but if you want muscle enhancement, you must have sufficient protein supply.
The eggs, milk and various meats on the table are rich in protein -rich foods. For humans, protein is available at hand; however, it is difficult to get the structure of the protein.

Diet rich in protein | Source: ISLide
Because the structure of the protein is very complicated: Simply put, amino acids form a protein, each of which has a structure link called the peptide bond, and its connection can form two different angles.
Then give you a simple mathematical problem: assuming 100 amino acids to form protein, then 99 peptide bonds are required, 99 peptide bonds have two different angles structures, and there will be three possible stable structures at different angles. That's the possibility of the 198th level of 38. If you are slowly poverty, it will not be completed from the universe to the present. This is the paradox of the Levin Sol -the protein structure is very very diverse. Essence
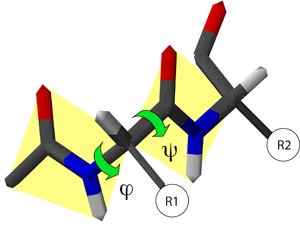
Different two -sided angle in the process of amino acid combination will generate different structures, so there is an infinite species structure like the Levv Solga paradox. Source: wikipedia
What should I do if the structure is so complicated? The most direct idea of biologists is observation, using different ways to measure: In the 1950s and 1960s, X -rays were used to diffuse — crystallizing protein, and then X -rays. Look, this difficulty is how to purify the protein.
Another popular research method is called freezing electron microscope, which is to use frozen slice technology and electronic microscope to directly see the protein structure, but the disadvantage is that it is very very expensive and burn money.
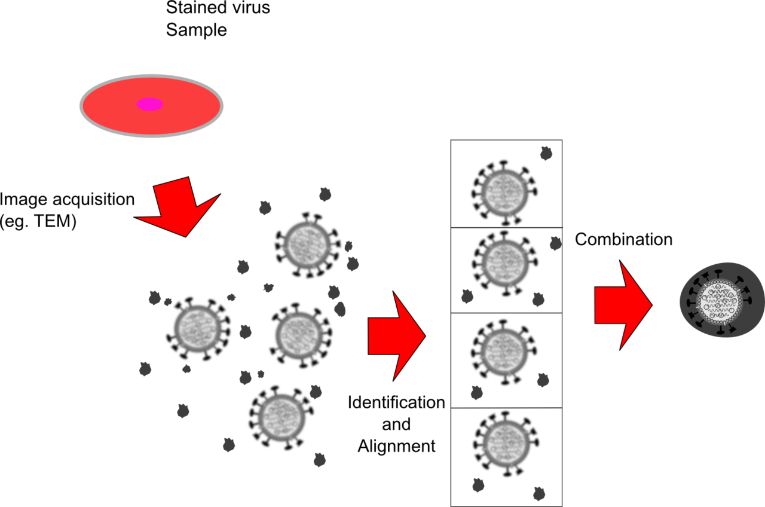
Basic principles of frozen electron microscope | Streaming: wikipedia
Then everyone guess so many methods, have been measured for decades, how many protein structures have we analyzed? In fact, there are a lot. According to the database records, the experiments have now analyzed 180,000 protein.
But in contrast, we just said that protein is composed of amino acids. As long as the sequencing technology is measured, the protein sequence can be derived. Finding the database can find that there are one billion protein sequences known now, which is nearly 10,000 times different from 180,000.
So structural biologists are very distressed -sequencing is too simple, resulting in the speed of structural biology far from keeping up.
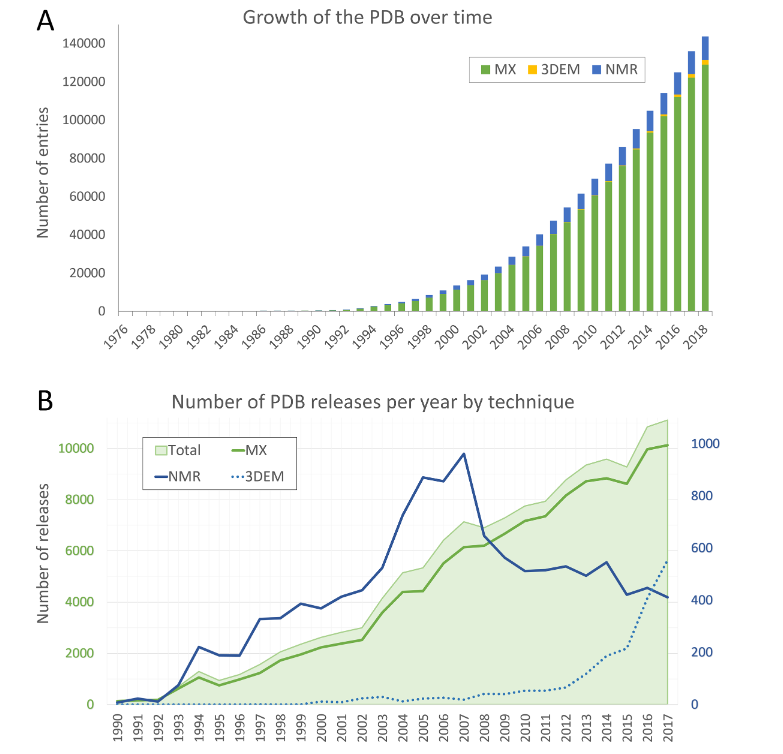
Verification of protein structure analysis changes | Pictorial source: nucleic acids research, 2019.
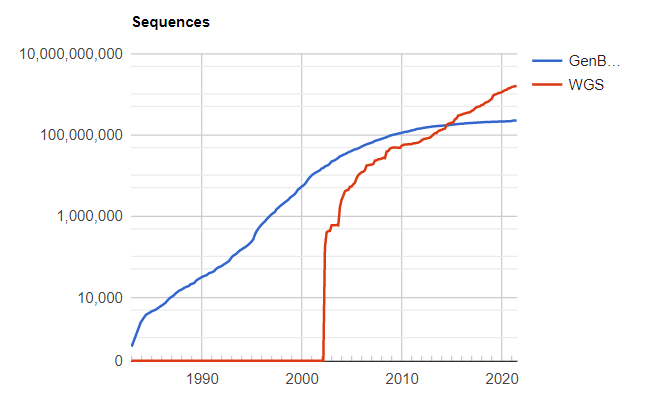
Speed of protein sequence sequencing | Power source: www.ncbi.nlm.nih.gov/genbank/statistics/
Therefore, many development algorithm computing biologists want to predict the structure, and it must be much faster than experiments through algorithm. But I just said that the paradox of Lvv Sol, the predicted structure is very, very difficult. If you are poor, it is an astronomical figure.
So there are many calculation ideas, such as I can analogize, I can analogize similar sequences in the experimental structure. It is speculated that similar sequences will have similar structures. This is called homologous modeling. After finishing the structure like the sewing, the structure is made up, this is called the threading method ... but so many methods have a problem: the accuracy is particularly poor. This is like I want to watch the high -definition video of 1080P, but there is only a mosaic high version of the mosaic. I can't see it clearly.

If you make an analogy, for example, the actual protein structure is the left, but the prediction results often only get the effect of the left figure, and many information cannot be known (only the signaling is processed. Exactly different differences) | Picture source: wikipedia
In order to promote the continuous movement of scientists in various countries, starting from 1994, CASP will be held every two years, called protein structure prediction key tests to evaluate everyone's algorithm predictions to improve the accuracy of the algorithm. Simply put, pick a few more from various protein sequences, and let the structural biologist do an experiment to solve a "standard answer", and then calculate the biologist to compare with his own algorithm to see who is the standard answer to the standard answer. Closer.
But unfortunately, 24 years have passed and still progress slowly.
Casp official website

Until 2018, a method called AlphaFold scored 80 points. Two years later, the second generation of Alphafold scored 90 points in 2020, which was basically the same as the standard answer made by the experiment. If you are just a metaphor of 1080P, others predict that it looks like a mosaic, but Alphafold2 forecast is almost 1000P, which is basically different from 1080P.
Everyone knows this is the artificial intelligence method developed by DeepMind. So last year's Top Ten Breakthroughs, in fact, there are also AI predictive protein structures.
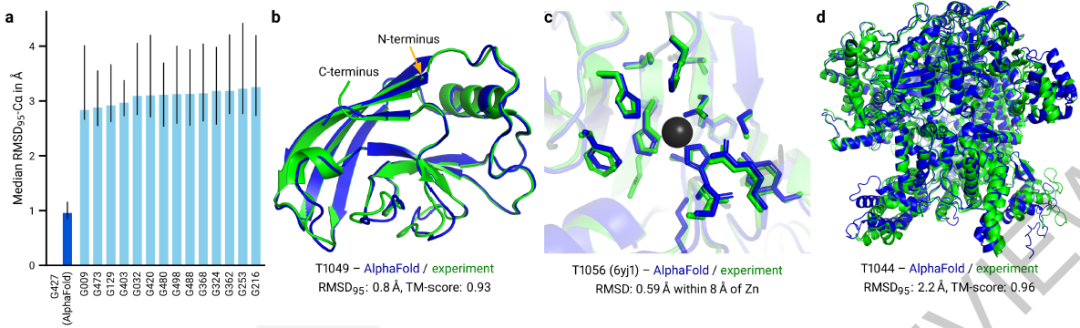
The accuracy of the Alphafold2 method predicted is far more than other algorithms (Figure A). At the same time, the predicted results and experimental results are basically consistent (Figure B-D) | Figure source: nature, 2021.
How can there be a breakthrough this year? This is because this algorithm is actually applied to biology.
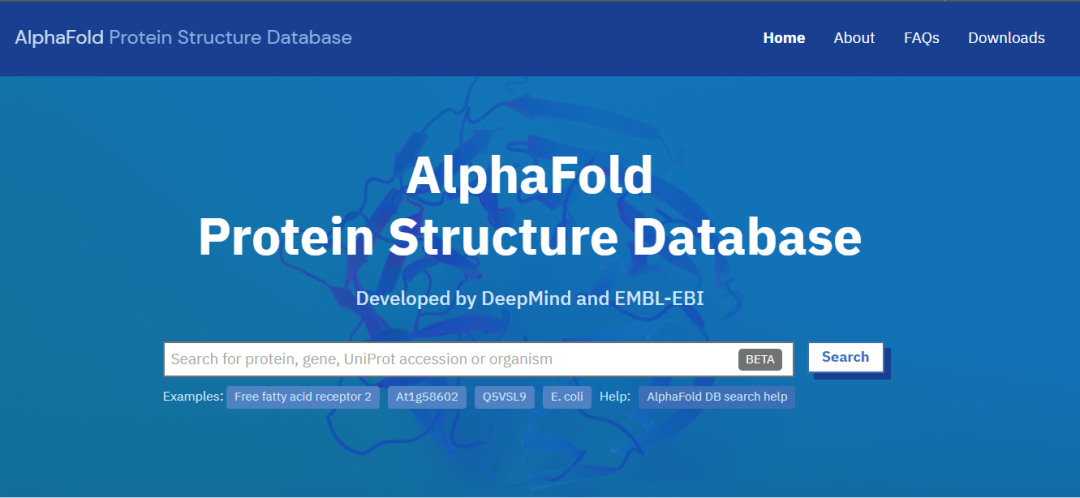
The first is the Alphafold2 algorithm developed by DeepMind. In just a few months, the decades of structured biologists analyzed the analysis of the unparalleled protein: 98% One -third can accurately predict, and some can also predict more than half. At the same time, they claimed that the database was extended to 100 million protein in the next few months. This is tens of times faster than the experimental method.
Protein structural database based on Alphafold2 predicted
Another biological application is RosettaFold, which is also developed based on artificial intelligence algorithms. It challenges more difficult areas -how to predict the combination of proteins and proteins, and also predicts the combination of thousands of proteins in a short time.
The most prominent promotional map of RosettaFold is the structural prediction of protein interaction

Many people may ask: What is the use of more than 100 million kinds of protein?
We initially mentioned that protein is everywhere in our lives, and protein has to play a function, the foundation is to have a certain structure. Therefore, predicting protein structure can help us better understand the function of protein, and then go to build protein molecular drugs, or study complex biochemical phenomena.
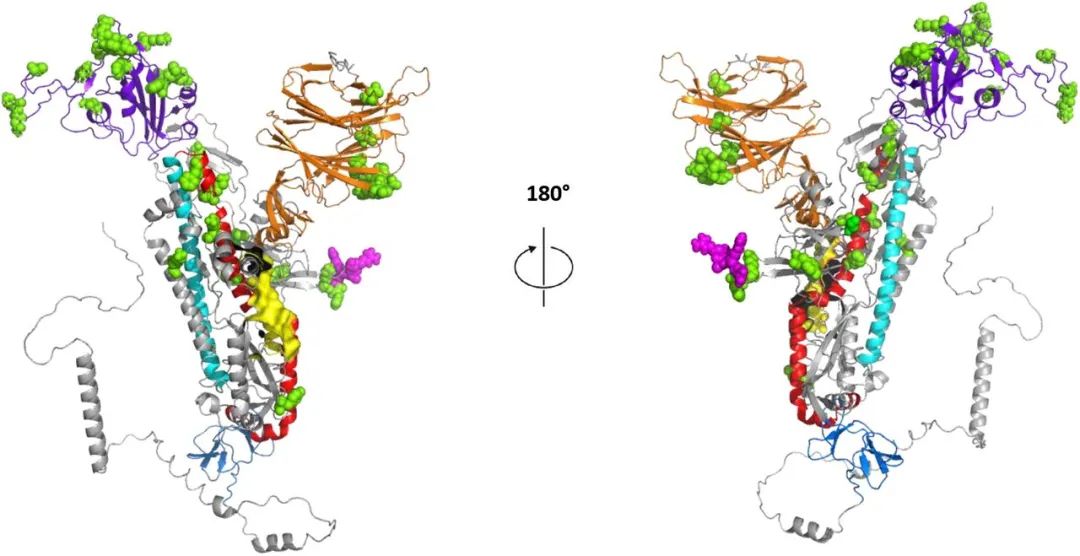
One of the simplest examples, for example, now we know that the new variation of the new coronary virus Omecko is particularly spread, and the key spiny protein structure of this dissemination can be predicted by artificial intelligence. Methods can be more effective for Omikon.
The S protein structure of the Omircong mutant prediction predicted by Alphafold
But at the same time, although it is said that artificial intelligence has completed a lot of structural biologists, this prediction is still incomplete:
For example, some complicated structures may not have been parsed in the experiment of structural biologists, and artificial intelligence has not been able to learn, so it is not predicted, so many problems still require in -depth inquiry of structural biologists;
There are also a lot of protein when they play a dynamic change process. The results of the prediction at this time are not accurate. If you use 1080P as an example, it is theoretically a 1080P video, but artificial intelligence predicts in these few seconds. It is 1080P HD, but it is predicted to be mosaic in those few seconds, so it is not accurate.
These are the flaws of artificial intelligence predicting protein, but the flaws are not covered. There are too many surprises that artificial intelligence in the predictive protein structure prediction, and this annual breakthrough is the best application for calculating science in life science.
Reprinted content only represents the author's point of view
Does not represent the position of the Institute of Physics of the Chinese Academy of Sciences
If you need to reprint, please contact the original public account
Source: BIOKIWI
Edit: Lezi Superman

- END -
Ningbo was selected as the nationwide recycled water use configuration pilot to take you to understand Ningbo's "regenerative water"

Recently, Ningbo has successfully selected the list of pilot cities in the first b...
New species!There are new discoveries in Yunnan again

recentlyIn Honghehani Hani and Yi Autonomous PrefectureJinping Miao Yao people and...