Progress | Study on a single molecule dynamic structure reveals the molecular mechanism of the key domain and DNA role of the pIF1 PIF1 PIF1
Author:Institute of Physics of the Ch Time:2022.07.29
Protein molecular machines play a vital role in life activities, and the structure in the process of exercise is highly dynamic. Traditional structural analysis methods have certain limitations for solving dynamic structures. Single molecular methods can realize the dynamic process of biomainer affected by the heating and rise in real time with high space -time resolution.
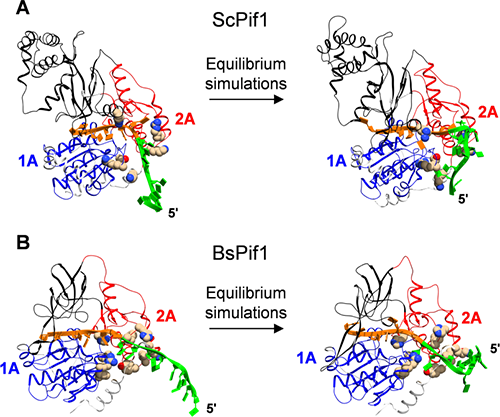
Figure 1 Molecular dynamic simulation reveals the combination of PIF1 to SSDNA
The Institute of Physics of the Chinese Academy of Sciences/Beijing Concrete Category Physics National Research Center SM4 Group SM4 Group SM4 has been committed to developing single molecular technology research biomolecular structure and its dynamics for many years. , NUCLEIC Acids Res. 2015, 43, 3736, Nucleic Acids Res. 2016, 44, 4330; Phys. Rev. Lett. 2017, 119, 138102; Nature2019, 567, 409; Nucleic Acids Res. 2020, 48, 3156; , 25, 103606) A series of results have been achieved in operating mechanism research. The main line of these studies is to obtain fine dynamic information of biomolecular molecules through high -precision single molecules. They developed DNA nano tensioners (NANOTENSIONER, PHYS. Rev. Lett. 2017), which can distinguish between 0.5 alkaline -based pairs, which is enough to clearly study the dynamic process of nucleic acid enzymes and the correlation between structure and function. Further application of high -spatial resolution DNA nano tensilers and other technologies, they and collaborators have studied monomer -of -the -ausprase enzymes (RECQ, PIF1) and six -yuan rotation enzyme (T7 GP4). , The regulation and control of the step -long force, the randomness of the steps, and the important role in the repair of DNA damage (phys. Rev. Lett. 2017, 119, 138102; j. Phys. Chem. B2018, 22, 5790; Nucleic Acids Res . 2020, 48, 3156). In another work, they and the collaborators studied the HH sub -turpans of BLM's dexterase and found that the sub -chain DNA had a dynamic interaction and had a variety of dynamic constructs. It is found that these interactions can regulate the composition of the complex and change its unproof mode, indicating that the importance of the sub -function in determining the BLM function and the possible molecular mechanism that causes Bloom syndrome (iScience2022, 25, 103606).
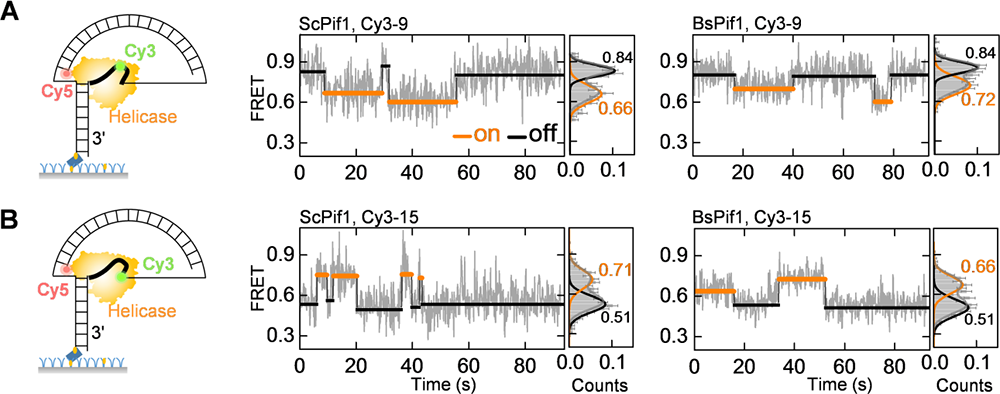
Figure 2 SMFRET enhanced by DNA nano tensioner reveals the dynamic bending conformity of DNA and PIF1
Recently, they and collaborators used single-molecule fluorescent resonance energy transfer, monoclonal magnetic mirror technology and molecular dynamic simulation, clarifying the dynamic interaction between the two RECA-Like sub-submalls of PIF1 aquarinase and DNA. Reca-Like sub-turks have sequence conservativeness and ATP binding sites, which is of great significance for PIF1 activity. However, the lack of structural information combined with these two subunits and DNA cannot explain the subtle but important differences of different dexterase in the PIF1 family. DNA nano tensioner has been further applied here. They found that, in addition to the 5-7 nucleotide combined with PIF1 in the crystal structure, the neighboring nucleotide dynamically combined on the surface of the PIF1 directly affects the process of PIF1 binding and unlocking DNA. Further, when this part of the nucleotide is combined on the surface of the enzyme, its structure is bent and dynamic, so the pulling effect of the tension and salt shielding can regulate the function of PIF1.
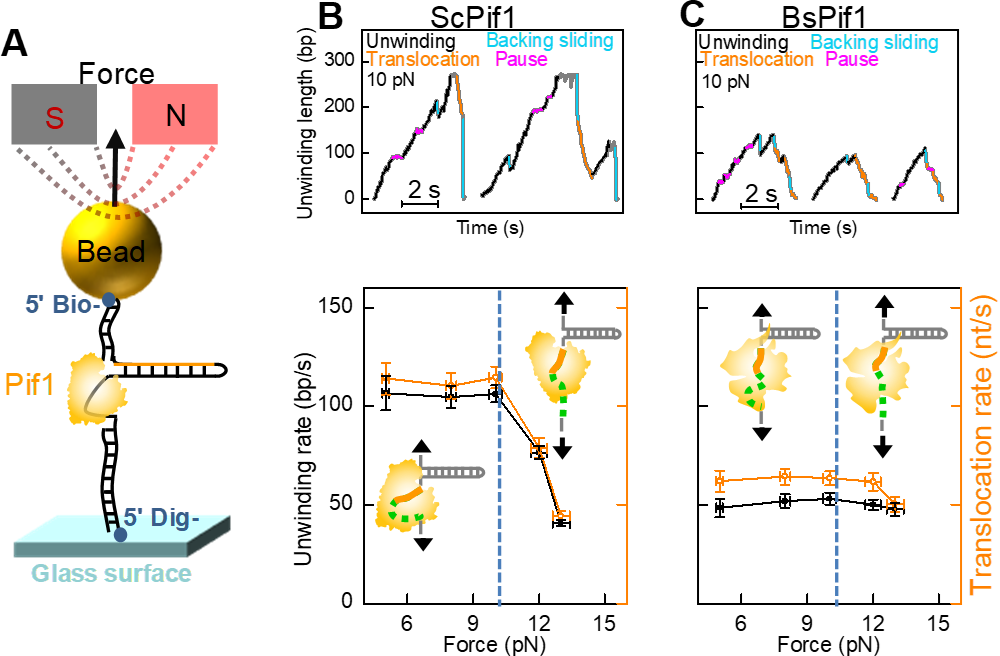
Figure 3 Single -molecular magnetic cymbals reveal the PIF1 unlocking and walking function of PIF1 under force adjustment
The work was published in the title of "Identification of Flexible Pif1 – DNA Interactions and their Impacts on Enzymatic Activities", published in the famous Biological Journal Nucleic Acids Research (if = 19.16, 2016), the Institute of Matheology of the Chinese Academy of Sciences Soft Research Institute Researcher and researcher at the Beijing Computing Science Research Center Liu Haiguang are the author's joint communication authors. Li Jinghua, a scientific assistant of physics institutes, postdoctoral Ma Jianbing, and Beijing Computing Science Research Center Vikash Kumar as the first author.
This work has received the National Natural Science Foundation of China (12090051, 11834018, 91753104, U1930402, 12090050, 1522409), the Chinese Academy of Sciences Pioneer B Project (XDB37000000), National Key R & D project (2019YFA0709304), and the Ministry of Science Study Plan for Frontier Science of the Chinese Academy of Sciences (ZDBS-LY-Slh015) and support of the Fund of Youth Innovation Promotion of the Chinese Academy of Sciences (Y2021003).Article link
edit:
- END -
The admission notice is delivered one after another. Are you get in these hidden functions?

At present, it is the admission of colleges and universities. Candidates who have ...
293,900 Yellow Stone people received interest on the provident fund
Dear ××, you 2021-2022 provident funds are at XX yuan, and your balance is currently ×× yuan. On the afternoon of June 30, many citizens successively received a settlement of a city provident fu